Dissociation PES curves with Qunova HiVQE
Usage estimate (NOTE: This is an estimate only. Your runtime may vary.)
- Li2S: 5 minutes QPU time on
ibm_marrakesh - FeP-NO: 5 minutes QPU time on
ibm_marrakesh
Background
Accurately calculating chemical reaction energies is crucial for scientific advancements in materials science, chemical engineering, drug discovery, and other fields. Among various chemical systems, the Li-S system has garnered significant interest for understanding and developing new battery compositions. This tutorial provides hands-on experience in calculating the Li-S bond dissociation potential energy surface (PES) of a system by removing one lithium atom using HiVQE calculations. The results can be compared with reference calculations (CASCI) as well as classical methods like Hartree-Fock (HF) for a 20-qubit problem.
Requirements
Install the following dependencies to run the code in this tutorial.
!pip install qiskit-ibm-catalog
!pip install qiskit_ibm_runtime
!pip install pyscf
!pip install numpy
!pip install matplotlibSetup
To run this tutorial, import qunova/hivqe-chemistry function via QiskitFunctionCatalog. You need an IBM Quantum® Premium Plan account with a license from Qunova to run this function.
from qiskit_ibm_catalog import QiskitFunctionsCatalog
from qiskit_ibm_runtime import QiskitRuntimeService
from pyscf import gto, scf, mcscf
import matplotlib.pyplot as plt
import pprint
service = QiskitRuntimeService()
catalog = QiskitFunctionsCatalog(
channel="ibm_cloud", instance="USER_CRN", token="USER_API_KEY"
)
hivqe = catalog.load("qunova/hivqe-chemistry")Part 1: Li2S (20Q)
Step 1: Map classical inputs to a quantum problem
Define geometries in dictionary format for different bond distance of Li-S to calculate PES curve. These geometries are optimized using B3LYP/631g calculations.
str_geometries = {
"1.51": "S -1.239044 0.671232 -0.030374; Li -1.506327 0.432403 -1.498949; Li -0.899996 0.973348 1.826768",
"1.91": "S -1.215858 0.692272 0.099232; Li -1.553305 0.390283 -1.758043; Li -0.876205 0.994426 1.956257",
"2.40": "S -1.741432 0.680397 0.346702; Li -0.529307 0.488006 -1.729343; Li -1.284307 0.989409 2.177209",
"3.10": "S -2.347450 0.657089 0.566194; Li -0.199353 0.527517 -1.665148; Li -1.008243 0.973206 1.893522",
"3.80": "S -2.707255 0.674298 0.909161; Li 0.079218 0.552012 -1.671656; Li -0.927010 0.931502 1.557063",
"4.50": "S -2.913363 0.709175 1.276987; Li 0.368656 0.559989 -1.798088; Li -1.010340 0.888647 1.315670",
}
str_geometriesOutput:
{'1.51': 'S -1.239044 0.671232 -0.030374; Li -1.506327 0.432403 -1.498949; Li -0.899996 0.973348 1.826768',
'1.91': 'S -1.215858 0.692272 0.099232; Li -1.553305 0.390283 -1.758043; Li -0.876205 0.994426 1.956257',
'2.40': 'S -1.741432 0.680397 0.346702; Li -0.529307 0.488006 -1.729343; Li -1.284307 0.989409 2.177209',
'3.10': 'S -2.347450 0.657089 0.566194; Li -0.199353 0.527517 -1.665148; Li -1.008243 0.973206 1.893522',
'3.80': 'S -2.707255 0.674298 0.909161; Li 0.079218 0.552012 -1.671656; Li -0.927010 0.931502 1.557063',
'4.50': 'S -2.913363 0.709175 1.276987; Li 0.368656 0.559989 -1.798088; Li -1.010340 0.888647 1.315670'}
HiVQE calculations will be performed with the options defined below. Using sto3g basis for , there are 19 spatial orbitals with 22 electrons. To run (10o,10e) case with HiVQE calculation, you can define 10 active orbitals and six frozen orbitals. At each iteration, 100 shots will be used to sample electron configuration generated by the ExcitationPreserving quantum circuit (epa) with circular entanglement and two repetitions (reps). The maximum number of iteration is set to be 30 to ensure termination of iteration with energy convergence.
molecule_options = {
"basis": "sto3g",
"active_orbitals": list(range(5, 15)),
"frozen_orbitals": list(range(5)),
}
hivqe_options = {
"shots": 100,
"max_iter": 30,
"ansatz": "epa",
"ansatz_entanglement": "circular",
"ansatz_reps": 2,
}Step 2 and 3: Optimize problem for quantum hardware execution and execute using the HiVQE Chemistry function
Set up the for loop to run HiVQE calculations with geometries with options defined below. Jobs are submitted in the for loop. In this tutorial, you will submit six geometries and retrieve results when they are all completed. In the main function run, you need to define the max_states and max_expansion_states to control the maximum size of the subspace matrix and to control how many states can be generated using classical CI expansion methods per iteration. The function job ids will be stored in the dictionary with each geometry label to further track and process the ouptut.
info_jobid = {}
for dis, geom in str_geometries.items():
hivqe_run = hivqe.run(
geometry=geom,
backend_name="",
max_states=40000,
max_expansion_states=100,
molecule_options=molecule_options,
hivqe_options=hivqe_options,
)
status = hivqe_run.status()
info_jobid[dis] = hivqe_run.job_id
print(info_jobid)Output:
{'1.51': 'de3b8818-c9db-4fa3-a3c2-d51551c2dfaf', '1.91': '55d9467a-fc85-49a8-9bc6-8f6990e421e5', '2.40': '415112b3-69ff-4d53-8b10-cb4e3be68c9e', '3.10': 'ef67b600-3887-4225-b872-e354dfdf8454', '3.80': 'b16d3502-a9e4-4560-9775-852e9d07e70f', '4.50': '0c0bffc7-af77-4a56-a656-2a2610c991d6'}
Let's check whether all jobs are still running or completed.
completed_jobs_num = 0
running_jobs_num = 0
completed_jobs = {}
for i, info in enumerate(info_jobid.items()):
dis, job_id = info
submitted_job = catalog.get_job_by_id(job_id)
stat = submitted_job.status()
print(dis, submitted_job.job_id, stat)
if stat == "DONE":
completed_jobs_num += 1
completed_jobs[dis] = submitted_job
if (stat == "RUNNING") or (stat == "QUEUED"):
running_jobs_num += 1
print(
f"Completed {completed_jobs_num} job, Running or Queued {running_jobs_num} job"
)Output:
1.51 de3b8818-c9db-4fa3-a3c2-d51551c2dfaf DONE
1.91 55d9467a-fc85-49a8-9bc6-8f6990e421e5 DONE
2.40 415112b3-69ff-4d53-8b10-cb4e3be68c9e DONE
3.10 ef67b600-3887-4225-b872-e354dfdf8454 DONE
3.80 b16d3502-a9e4-4560-9775-852e9d07e70f DONE
4.50 0c0bffc7-af77-4a56-a656-2a2610c991d6 DONE
Completed 6 job, Running or Queued 0 job
Once all jobs are completed, let's retrieve all calculation results.
hivqe_result = {}
if len(info_jobid) == completed_jobs_num:
print("All jobs are completed")
for i, job in enumerate(completed_jobs.items()):
dis, cal = job
print(dis, cal.result()["energy"])
hivqe_result[str(dis)] = cal.result()["energy"]Output:
All jobs are completed
1.51 -407.8944801731773
1.91 -407.9800570932916
2.40 -407.9372992999806
3.10 -407.86278336000134
3.80 -407.83092972296157
4.50 -407.82971011225766
pprint.pprint(hivqe_result)Output:
{'1.51': -407.8944801731773,
'1.91': -407.9800570932916,
'2.40': -407.9372992999806,
'3.10': -407.86278336000134,
'3.80': -407.83092972296157,
'4.50': -407.82971011225766}
The entire QPU runtime used in the job can be tracked by logging in to quantum.ibm.com and viewing submitted jobs with the qunova-chemistry-hivqe tag.
Step 4: Post-process and compare with classical methods
Classical reference calculation (CASCI) can be conducted for (10o,10e) to validate HiVQE results.
str_geometries = {
"1.31": "S -1.250686 0.660708 -0.095168; Li -1.482812 0.453464 -1.369406; Li -0.911870 0.962810 1.762020",
"1.41": "S -1.244856 0.665971 -0.062773; Li -1.494574 0.442933 -1.434177; Li -0.905937 0.968078 1.794395",
"1.51": "S -1.239044 0.671232 -0.030374; Li -1.506327 0.432403 -1.498949; Li -0.899996 0.973348 1.826768",
"1.61": "S -1.233245 0.676492 0.002027; Li -1.518073 0.421873 -1.563722; Li -0.894049 0.978617 1.859141",
"1.71": "S -1.227453 0.681752 0.034429; Li -1.529816 0.411343 -1.628496; Li -0.888099 0.983887 1.891513",
"1.81": "S -1.221659 0.687012 0.066831; Li -1.541558 0.400813 -1.693270; Li -0.882150 0.989157 1.923885",
"1.91": "S -1.215858 0.692272 0.099232; Li -1.553305 0.390283 -1.758043; Li -0.876205 0.994426 1.956257",
"2.01": "S -1.209887 0.697544 0.131599; Li -1.565136 0.379748 -1.822800; Li -0.870344 0.999691 1.988646",
"2.11": "S -1.203945 0.702813 0.163973; Li -1.576953 0.369214 -1.887560; Li -0.864469 1.004956 2.021033",
"2.21": "S -1.198023 0.708081 0.196350; Li -1.588760 0.358680 -1.952322; Li -0.858584 1.010221 2.053417",
"2.30": "S -1.365426 0.717714 0.367060; Li -0.689401 0.458925 -1.828368; Li -1.500219 0.981173 2.255876",
"2.31": "S -1.192118 0.713348 0.228731; Li -1.600559 0.348146 -2.017085; Li -0.852690 1.015488 2.085800",
"2.40": "S -1.741432 0.680397 0.346702; Li -0.529307 0.488006 -1.729343; Li -1.284307 0.989409 2.177209",
"2.50": "S -1.885961 0.669986 0.365815; Li -0.461563 0.499084 -1.695846; Li -1.207523 0.988741 2.124599",
"2.60": "S -1.977163 0.665155 0.389784; Li -0.416654 0.504966 -1.683655; Li -1.161229 0.987690 2.088439",
"2.70": "S -2.063642 0.661518 0.418977; Li -0.367600 0.510505 -1.676408; Li -1.123804 0.985788 2.051998",
"2.80": "S -2.141072 0.659218 0.451663; Li -0.323153 0.515056 -1.673046; Li -1.090821 0.983538 2.015951",
"2.90": "S -2.212097 0.657968 0.487535; Li -0.281989 0.518909 -1.672407; Li -1.060960 0.980935 1.979440",
"3.00": "S -2.281477 0.657123 0.525155; Li -0.239607 0.523326 -1.668669; Li -1.033963 0.977363 1.938081",
"3.10": "S -2.347450 0.657089 0.566194; Li -0.199353 0.527517 -1.665148; Li -1.008243 0.973206 1.893522",
"3.20": "S -2.410882 0.657532 0.608912; Li -0.157788 0.532069 -1.659971; Li -0.986376 0.968211 1.845627",
"3.30": "S -2.470306 0.658818 0.654893; Li -0.118007 0.536237 -1.656311; Li -0.966733 0.962757 1.795986",
"3.40": "S -2.525776 0.660762 0.702910; Li -0.078312 0.540189 -1.654076; Li -0.950958 0.956861 1.745734",
"3.50": "S -2.576885 0.663376 0.752788; Li -0.039076 0.543706 -1.654536; Li -0.939085 0.950730 1.696316",
"3.60": "S -2.623930 0.666534 0.803853; Li 0.000274 0.546839 -1.657697; Li -0.931390 0.944439 1.648412",
"3.70": "S -2.667364 0.670217 0.856250; Li 0.039572 0.549616 -1.663265; Li -0.927254 0.937980 1.601583",
"3.80": "S -2.707255 0.674298 0.909161; Li 0.079218 0.552012 -1.671656; Li -0.927010 0.931502 1.557063",
"3.90": "S -2.744005 0.678718 0.962425; Li 0.119268 0.554073 -1.682595; Li -0.930310 0.925021 1.514738",
"4.00": "S -2.777891 0.683415 1.015798; Li 0.159751 0.555810 -1.696024; Li -0.936907 0.918587 1.474794",
"4.10": "S -2.809179 0.688333 1.069057; Li 0.200678 0.557234 -1.711873; Li -0.946546 0.912245 1.437385",
"4.20": "S -2.838194 0.693443 1.122205; Li 0.242066 0.558401 -1.729770; Li -0.958918 0.905968 1.402134",
"4.30": "S -2.864984 0.698619 1.174415; Li 0.283858 0.559186 -1.750539; Li -0.973920 0.900007 1.370693",
"4.40": "S -2.889984 0.703887 1.226140; Li 0.326068 0.559728 -1.773231; Li -0.991131 0.894196 1.341660",
"4.50": "S -2.913363 0.709175 1.276987; Li 0.368656 0.559989 -1.798088; Li -1.010340 0.888647 1.315670",
}rhf_result = {}
casci_result = {}
cas_list = molecule_options["active_orbitals"]
distance_ref = []
for dis, geom in str_geometries.items():
distance_ref.append(dis)
mole = gto.M(atom=geom, basis=molecule_options["basis"])
mole.verbose = 0
# RHF energy
mf = scf.RHF(mole).run()
mo_occ = mf.mo_occ
num_elecs_as = int(sum([mo_occ[idx] for idx in cas_list]))
rhf_result[str(dis)] = mf.e_tot
# CASCI energy
casci_solver = mcscf.CASCI(mf, len(cas_list), num_elecs_as)
orbs = mcscf.addons.sort_mo(casci_solver, mf.mo_coeff, cas_list, base=0)
casci_solver.kernel(orbs)
casci_result[str(dis)] = casci_solver.e_tot
print(
f"d={dis:4.3} RHF Energy: {mf.e_tot:14.10}, CASCI Energy: {casci_solver.e_tot:14.10}"
)Output:
d=1.3 RHF Energy: -407.7137006, CASCI Energy: -407.7193917
d=1.4 RHF Energy: -407.8183196, CASCI Energy: -407.8245211
d=1.5 RHF Energy: -407.8878013, CASCI Energy: -407.8944802
d=1.6 RHF Energy: -407.9315356, CASCI Energy: -407.9385663
d=1.7 RHF Energy: -407.9569034, CASCI Energy: -407.9641258
d=1.8 RHF Energy: -407.9693681, CASCI Energy: -407.9766313
d=1.9 RHF Energy: -407.9728592, CASCI Energy: -407.9800572
d=2.0 RHF Energy: -407.9701684, CASCI Energy: -407.9772549
d=2.1 RHF Energy: -407.9632701, CASCI Energy: -407.9702381
d=2.2 RHF Energy: -407.9535584, CASCI Energy: -407.9604007
d=2.3 RHF Energy: -407.9420173, CASCI Energy: -407.9487043
d=2.3 RHF Energy: -407.9420156, CASCI Energy: -407.9487024
d=2.4 RHF Energy: -407.9297216, CASCI Energy: -407.9372993
d=2.5 RHF Energy: -407.9172, CASCI Energy: -407.9261859
d=2.6 RHF Energy: -407.9061139, CASCI Energy: -407.915961
d=2.7 RHF Energy: -407.8937118, CASCI Energy: -407.904259
d=2.8 RHF Energy: -407.8816389, CASCI Energy: -407.8928292
d=2.9 RHF Energy: -407.8700448, CASCI Energy: -407.8819574
d=3.0 RHF Energy: -407.859054, CASCI Energy: -407.8719092
d=3.1 RHF Energy: -407.8487619, CASCI Energy: -407.8628304
d=3.2 RHF Energy: -407.8392304, CASCI Energy: -407.8548482
d=3.3 RHF Energy: -407.8304842, CASCI Energy: -407.8480217
d=3.4 RHF Energy: -407.8225124, CASCI Energy: -407.8423743
d=3.5 RHF Energy: -407.8152758, CASCI Energy: -407.8378892
d=3.6 RHF Energy: -407.8087161, CASCI Energy: -407.8345331
d=3.7 RHF Energy: -407.802764, CASCI Energy: -407.8322563
d=3.8 RHF Energy: -407.7973458, CASCI Energy: -407.83093
d=3.9 RHF Energy: -407.7923883, CASCI Energy: -407.8303555
d=4.0 RHF Energy: -407.7878216, CASCI Energy: -407.83025
d=4.1 RHF Energy: -407.783582, CASCI Energy: -407.8303243
d=4.2 RHF Energy: -407.7796124, CASCI Energy: -407.8303791
d=4.3 RHF Energy: -407.7758633, CASCI Energy: -407.8302885
d=4.4 RHF Energy: -407.7722923, CASCI Energy: -407.8300614
d=4.5 RHF Energy: -407.7688641, CASCI Energy: -407.829711
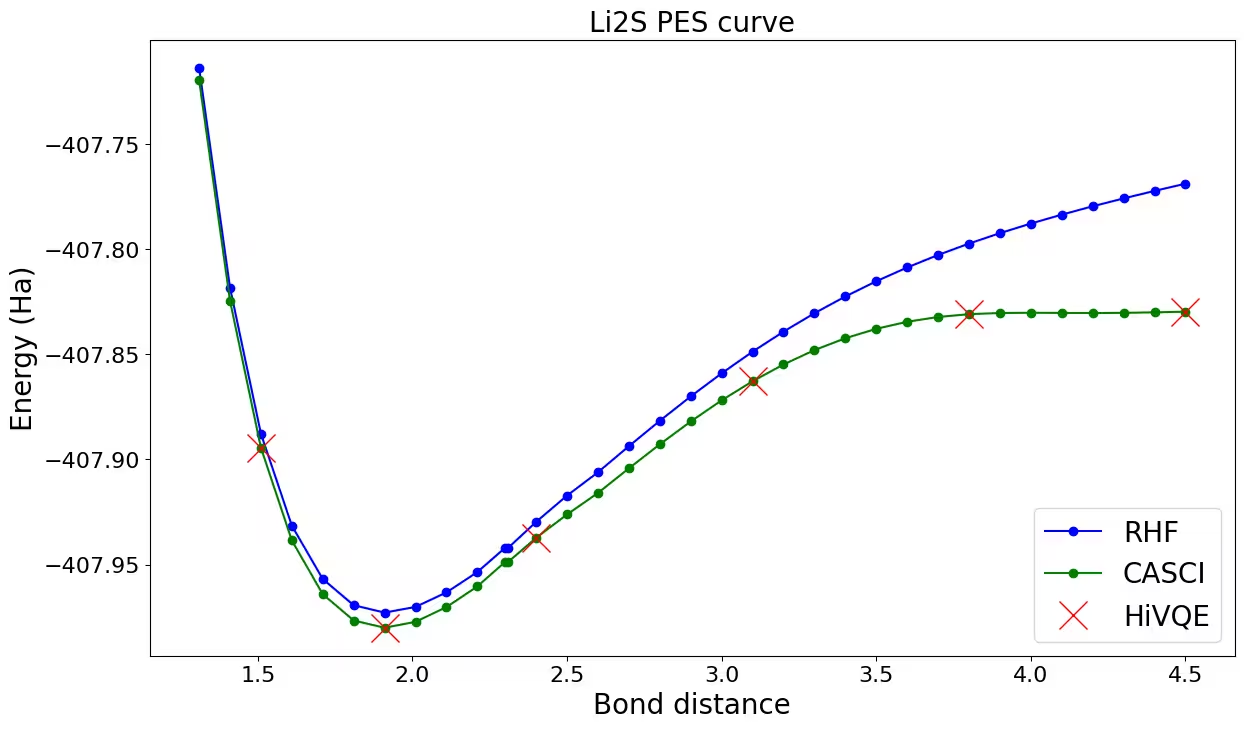
Plotting the dissociation curve for Li_2S
Let's plot and compare HiVQE results with HF and CASCI. You can observe that all HiVQE calculations are well-matched with the classical reference result (CASCI).
fig, ax = plt.subplots(1, 1)
hf_energy = [v for key, v in rhf_result.items()]
casci_energy = [v for key, v in casci_result.items()]
hivqe_energy = [v for key, v in hivqe_result.items()]
distance_ref = [float(key) for key, v in rhf_result.items()]
distance = [float(key) for key, v in hivqe_result.items()]
ax.plot(distance_ref, hf_energy, "-o", label="RHF", c="blue")
ax.plot(distance_ref, casci_energy, "-o", label="CASCI", c="green")
ax.plot(distance, hivqe_energy, "x", label="HiVQE", c="red", markersize=20)
ax.legend(fontsize=20)
ax.tick_params("both", labelsize=16)
ax.set_xlabel("Bond distance (angstrom)", size=20)
ax.set_ylabel("Energy (Ha)", size=20)
ax.set_title("Li2S PES curve", size=20)
fig.set_size_inches(14, 8)Output:
Part 2: FeP-NO (44Q)
Step 1: Map classical inputs to a quantum problem
Define the options for HiVQE calculations
molecule_options = {
"basis": "631g*",
"active_orbitals": list(range(90, 112, 1)),
"frozen_orbitals": list(range(0, 90, 1)),
"charge": -1,
}
hivqe_options = {
"shots": 2000,
"max_iter": 40,
"ansatz": "epa",
"ansatz_entanglement": "linear",
"ansatz_reps": 2,
"amplitude_screening_tolerance": 1e-6,
}Define FeP-NO geometries in dictionary format for different bond distances of Fe-N to calculate PES curve.
geometry_1_75 = """
Fe 9.910596 31.534095 1.798088
N 10.557481 31.888419 -0.055204
N 11.823496 31.255002 2.384659
N 9.292831 30.783362 3.568730
N 8.036805 31.418327 1.124265
C 9.784765 32.177349 -1.158798
C 10.612656 32.501029 -2.296868
C 11.903375 32.404043 -1.876832
C 11.859093 32.028943 -0.483750
C 12.965737 31.464698 1.641427
C 14.146517 31.236323 2.440231
C 13.713061 30.885870 3.681911
C 12.268752 30.896411 3.634891
C 10.067717 30.486167 4.664747
C 9.246224 30.053411 5.772052
C 7.957075 30.082846 5.336488
C 7.995710 30.538421 3.967046
C 6.900258 31.104497 1.836595
C 5.722470 31.251707 1.015333
C 6.148430 31.668586 -0.207993
C 7.587039 31.767438 -0.130483
C 8.399453 32.134197 -1.192329
H 7.912872 32.388031 -2.131079
C 12.984883 31.836053 0.306093
H 13.955948 31.977044 -0.162626
C 11.453768 30.560663 4.708020
H 11.940677 30.298823 5.644352
C 6.877071 30.697580 3.164102
H 5.907240 30.476797 3.603674
H 12.813946 32.569160 -2.441577
H 10.236332 32.758110 -3.280309
H 15.164312 31.335191 2.080201
H 14.299625 30.629109 4.556760
H 9.626524 29.758225 6.743433
H 7.053076 29.823583 5.875809
H 4.709768 31.058315 1.350561
H 5.561898 31.886355 -1.093106
N 9.832739 33.209042 2.298783
O 9.346337 34.075996 1.606023
"""
geometry_2_00 = """
Fe 9.917990 31.445558 1.778346
N 10.556809 31.866188 -0.055498
N 11.814089 31.227003 2.372666
N 9.297875 30.758246 3.550104
N 8.043584 31.397768 1.120485
C 9.784831 32.164652 -1.160219
C 10.611624 32.501801 -2.293514
C 11.902858 32.406547 -1.875160
C 11.859552 32.017818 -0.486307
C 12.960503 31.454432 1.636717
C 14.140770 31.242960 2.439615
C 13.708543 30.884151 3.678983
C 12.266351 30.874173 3.627468
C 10.070264 30.465070 4.655102
C 9.247247 30.053101 5.766681
C 7.958085 30.091201 5.332866
C 7.998432 30.529979 3.958727
C 6.901428 31.093932 1.833807
C 5.723289 31.255057 1.016540
C 6.151314 31.670649 -0.206350
C 7.589736 31.755538 -0.133074
C 8.400230 32.124963 -1.194447
H 7.913264 32.386655 -2.130914
C 12.983905 31.827747 0.302415
H 13.955696 31.979687 -0.161365
C 11.454251 30.533644 4.698234
H 11.941002 30.276716 5.636156
C 6.877444 30.689985 3.159940
H 5.907605 30.480118 3.604825
H 12.813105 32.581608 -2.437367
H 10.233725 32.768337 -3.273979
H 15.157796 31.357524 2.082132
H 14.295001 30.638320 4.557047
H 9.626721 29.768762 6.741623
H 7.051752 29.847502 5.875478
H 4.709710 31.071712 1.354640
H 5.565103 31.898376 -1.089333
N 9.840508 33.353531 2.373019
O 9.344561 34.158205 1.637232
"""
geometry_5_00 = """
Fe 9.918629 31.289202 1.717339
N 10.542914 31.832173 -0.080685
N 11.795572 31.199413 2.341831
N 9.294593 30.741247 3.513929
N 8.042689 31.359481 1.087282
C 9.775254 32.111817 -1.200449
C 10.600219 32.479101 -2.319680
C 11.891090 32.425876 -1.887580
C 11.847694 32.024341 -0.507342
C 12.945734 31.464689 1.611366
C 14.116395 31.289997 2.423572
C 13.685777 30.915122 3.663719
C 12.252381 30.861042 3.608186
C 10.062170 30.463021 4.634102
C 9.236749 30.104333 5.755782
C 7.945687 30.161198 5.324720
C 7.989641 30.552269 3.941498
C 6.892881 31.087489 1.815829
C 5.722676 31.253502 1.001149
C 6.153153 31.631057 -0.238233
C 7.586010 31.695401 -0.179773
C 8.390724 32.047572 -1.247553
H 7.903308 32.291586 -2.187969
C 12.973334 31.849872 0.283741
H 13.944682 32.031190 -0.169145
C 11.447158 30.518591 4.678739
H 11.934423 30.277429 5.619969
C 6.864795 30.711643 3.146118
H 5.893357 30.532078 3.599511
H 12.800139 32.636412 -2.439296
H 10.224017 32.743662 -3.301293
H 15.131785 31.441247 2.076257
H 14.273933 30.694315 4.546802
H 9.612512 29.848040 6.739754
H 7.036117 29.960530 5.879248
H 4.707408 31.099933 1.347803
H 5.564992 31.851940 -1.121294
N 9.666041 36.091609 3.085945
O 9.598728 37.226756 3.411299
"""
str_geometries = {
"1.75": geometry_1_75,
"2.00": geometry_2_00,
"5.00": geometry_5_00,
}
hivqe_result = {}Output:
{'5.0': '\nFe 9.918629 31.289202 1.717339\nN 10.542914 31.832173 -0.080685\nN 11.795572 31.199413 2.341831\nN 9.294593 30.741247 3.513929\nN 8.042689 31.359481 1.087282\nC 9.775254 32.111817 -1.200449\nC 10.600219 32.479101 -2.319680\nC 11.891090 32.425876 -1.887580\nC 11.847694 32.024341 -0.507342\nC 12.945734 31.464689 1.611366\nC 14.116395 31.289997 2.423572\nC 13.685777 30.915122 3.663719\nC 12.252381 30.861042 3.608186\nC 10.062170 30.463021 4.634102\nC 9.236749 30.104333 5.755782\nC 7.945687 30.161198 5.324720\nC 7.989641 30.552269 3.941498\nC 6.892881 31.087489 1.815829\nC 5.722676 31.253502 1.001149\nC 6.153153 31.631057 -0.238233\nC 7.586010 31.695401 -0.179773\nC 8.390724 32.047572 -1.247553\nH 7.903308 32.291586 -2.187969\nC 12.973334 31.849872 0.283741\nH 13.944682 32.031190 -0.169145\nC 11.447158 30.518591 4.678739\nH 11.934423 30.277429 5.619969\nC 6.864795 30.711643 3.146118\nH 5.893357 30.532078 3.599511\nH 12.800139 32.636412 -2.439296\nH 10.224017 32.743662 -3.301293\nH 15.131785 31.441247 2.076257\nH 14.273933 30.694315 4.546802\nH 9.612512 29.848040 6.739754\nH 7.036117 29.960530 5.879248\nH 4.707408 31.099933 1.347803\nH 5.564992 31.851940 -1.121294\nN 9.666041 36.091609 3.085945\nO 9.598728 37.226756 3.411299\n'}
geometry_1_75 = """
Fe 9.910596 31.534095 1.798088
N 10.557481 31.888419 -0.055204
N 11.823496 31.255002 2.384659
N 9.292831 30.783362 3.568730
N 8.036805 31.418327 1.124265
C 9.784765 32.177349 -1.158798
C 10.612656 32.501029 -2.296868
C 11.903375 32.404043 -1.876832
C 11.859093 32.028943 -0.483750
C 12.965737 31.464698 1.641427
C 14.146517 31.236323 2.440231
C 13.713061 30.885870 3.681911
C 12.268752 30.896411 3.634891
C 10.067717 30.486167 4.664747
C 9.246224 30.053411 5.772052
C 7.957075 30.082846 5.336488
C 7.995710 30.538421 3.967046
C 6.900258 31.104497 1.836595
C 5.722470 31.251707 1.015333
C 6.148430 31.668586 -0.207993
C 7.587039 31.767438 -0.130483
C 8.399453 32.134197 -1.192329
H 7.912872 32.388031 -2.131079
C 12.984883 31.836053 0.306093
H 13.955948 31.977044 -0.162626
C 11.453768 30.560663 4.708020
H 11.940677 30.298823 5.644352
C 6.877071 30.697580 3.164102
H 5.907240 30.476797 3.603674
H 12.813946 32.569160 -2.441577
H 10.236332 32.758110 -3.280309
H 15.164312 31.335191 2.080201
H 14.299625 30.629109 4.556760
H 9.626524 29.758225 6.743433
H 7.053076 29.823583 5.875809
H 4.709768 31.058315 1.350561
H 5.561898 31.886355 -1.093106
N 9.832739 33.209042 2.298783
O 9.346337 34.075996 1.606023
"""
geometry_2_00 = """
Fe 9.917990 31.445558 1.778346
N 10.556809 31.866188 -0.055498
N 11.814089 31.227003 2.372666
N 9.297875 30.758246 3.550104
N 8.043584 31.397768 1.120485
C 9.784831 32.164652 -1.160219
C 10.611624 32.501801 -2.293514
C 11.902858 32.406547 -1.875160
C 11.859552 32.017818 -0.486307
C 12.960503 31.454432 1.636717
C 14.140770 31.242960 2.439615
C 13.708543 30.884151 3.678983
C 12.266351 30.874173 3.627468
C 10.070264 30.465070 4.655102
C 9.247247 30.053101 5.766681
C 7.958085 30.091201 5.332866
C 7.998432 30.529979 3.958727
C 6.901428 31.093932 1.833807
C 5.723289 31.255057 1.016540
C 6.151314 31.670649 -0.206350
C 7.589736 31.755538 -0.133074
C 8.400230 32.124963 -1.194447
H 7.913264 32.386655 -2.130914
C 12.983905 31.827747 0.302415
H 13.955696 31.979687 -0.161365
C 11.454251 30.533644 4.698234
H 11.941002 30.276716 5.636156
C 6.877444 30.689985 3.159940
H 5.907605 30.480118 3.604825
H 12.813105 32.581608 -2.437367
H 10.233725 32.768337 -3.273979
H 15.157796 31.357524 2.082132
H 14.295001 30.638320 4.557047
H 9.626721 29.768762 6.741623
H 7.051752 29.847502 5.875478
H 4.709710 31.071712 1.354640
H 5.565103 31.898376 -1.089333
N 9.840508 33.353531 2.373019
O 9.344561 34.158205 1.637232
"""
geometry_5_00 = """
Fe 9.918629 31.289202 1.717339
N 10.542914 31.832173 -0.080685
N 11.795572 31.199413 2.341831
N 9.294593 30.741247 3.513929
N 8.042689 31.359481 1.087282
C 9.775254 32.111817 -1.200449
C 10.600219 32.479101 -2.319680
C 11.891090 32.425876 -1.887580
C 11.847694 32.024341 -0.507342
C 12.945734 31.464689 1.611366
C 14.116395 31.289997 2.423572
C 13.685777 30.915122 3.663719
C 12.252381 30.861042 3.608186
C 10.062170 30.463021 4.634102
C 9.236749 30.104333 5.755782
C 7.945687 30.161198 5.324720
C 7.989641 30.552269 3.941498
C 6.892881 31.087489 1.815829
C 5.722676 31.253502 1.001149
C 6.153153 31.631057 -0.238233
C 7.586010 31.695401 -0.179773
C 8.390724 32.047572 -1.247553
H 7.903308 32.291586 -2.187969
C 12.973334 31.849872 0.283741
H 13.944682 32.031190 -0.169145
C 11.447158 30.518591 4.678739
H 11.934423 30.277429 5.619969
C 6.864795 30.711643 3.146118
H 5.893357 30.532078 3.599511
H 12.800139 32.636412 -2.439296
H 10.224017 32.743662 -3.301293
H 15.131785 31.441247 2.076257
H 14.273933 30.694315 4.546802
H 9.612512 29.848040 6.739754
H 7.036117 29.960530 5.879248
H 4.707408 31.099933 1.347803
H 5.564992 31.851940 -1.121294
N 9.666041 36.091609 3.085945
O 9.598728 37.226756 3.411299
"""
str_geometries = {
"1.75": geometry_1_75,
"2.00": geometry_2_00,
"5.00": geometry_5_00,
}
hivqe_result = {}Output:
{'5.0': '\nFe 9.918629 31.289202 1.717339\nN 10.542914 31.832173 -0.080685\nN 11.795572 31.199413 2.341831\nN 9.294593 30.741247 3.513929\nN 8.042689 31.359481 1.087282\nC 9.775254 32.111817 -1.200449\nC 10.600219 32.479101 -2.319680\nC 11.891090 32.425876 -1.887580\nC 11.847694 32.024341 -0.507342\nC 12.945734 31.464689 1.611366\nC 14.116395 31.289997 2.423572\nC 13.685777 30.915122 3.663719\nC 12.252381 30.861042 3.608186\nC 10.062170 30.463021 4.634102\nC 9.236749 30.104333 5.755782\nC 7.945687 30.161198 5.324720\nC 7.989641 30.552269 3.941498\nC 6.892881 31.087489 1.815829\nC 5.722676 31.253502 1.001149\nC 6.153153 31.631057 -0.238233\nC 7.586010 31.695401 -0.179773\nC 8.390724 32.047572 -1.247553\nH 7.903308 32.291586 -2.187969\nC 12.973334 31.849872 0.283741\nH 13.944682 32.031190 -0.169145\nC 11.447158 30.518591 4.678739\nH 11.934423 30.277429 5.619969\nC 6.864795 30.711643 3.146118\nH 5.893357 30.532078 3.599511\nH 12.800139 32.636412 -2.439296\nH 10.224017 32.743662 -3.301293\nH 15.131785 31.441247 2.076257\nH 14.273933 30.694315 4.546802\nH 9.612512 29.848040 6.739754\nH 7.036117 29.960530 5.879248\nH 4.707408 31.099933 1.347803\nH 5.564992 31.851940 -1.121294\nN 9.666041 36.091609 3.085945\nO 9.598728 37.226756 3.411299\n'}
Step 2 and 3: Optimize problem for quantum hardware execution and execute using the HiVQE Chemistry function
Based on the setup of HiVQE and geometries, obtain results sequentially.
Submit d(Fe-N) = 1.75 calculation.
hivqe_run_1_75 = hivqe.run(
geometry=str_geometries["1.75"],
backend_name="",
max_states=400000000,
max_expansion_states=100,
molecule_options=molecule_options,
hivqe_options=hivqe_options,
)
info_jobid_1_75 = hivqe_run_1_75.job_idTrack the job and retrieve the result for d(Fe-N) = 1.75 calculation.
submitted_job_1_75 = catalog.get_job_by_id(info_jobid_1_75)
stat = submitted_job_1_75.status()
print(submitted_job_1_75.job_id, stat)
if stat == "DONE":
hivqe_run_1_75_energy = submitted_job_1_75.result()["energy"]
print(f"Completed HiVQE calculation, Energy {hivqe_run_1_75_energy}")
hivqe_result["1.75"] = hivqe_run_1_75_energySubmit d(Fe-N) = 2.00 calculation.
hivqe_run_2_00 = hivqe.run(
geometry=str_geometries["2.00"],
backend_name="",
max_states=400000000,
max_expansion_states=100,
molecule_options=molecule_options,
hivqe_options=hivqe_options,
)
info_jobid_2_00 = hivqe_run_2_00.job_idTrack the job and retrieve the result for d(Fe-N) = 2.00 calculation.
submitted_job_2_00 = catalog.get_job_by_id(info_jobid_2_00)
stat = submitted_job_2_00.status()
print(submitted_job_2_00.job_id, stat)
if stat == "DONE":
hivqe_run_2_00_energy = submitted_job_2_00.result()["energy"]
print(f"Completed HiVQE calculation, Energy {hivqe_run_2_00_energy}")
hivqe_result["2.00"] = hivqe_run_2_00_energySubmit d(Fe-N) = 5.00 calculation.
hivqe_run_5_00 = hivqe.run(
geometry=str_geometries["5.00"],
backend_name="",
max_states=400000000,
max_expansion_states=100,
molecule_options=molecule_options,
hivqe_options=hivqe_options,
)
info_jobid_5_00 = hivqe_run_5_00.job_idTrack the job and retrieve the result for d(Fe-N) = 5.00 calculation.
submitted_job_5_00 = catalog.get_job_by_id(info_jobid_5_00)
stat = submitted_job_5_00.status()
print(submitted_job_5_00.job_id, stat)
if stat == "DONE":
hivqe_run_5_00_energy = submitted_job_5_00.result()["energy"]
print(f"Completed HiVQE calculation, Energy {hivqe_run_5_00_energy}")
hivqe_result["5.00"] = hivqe_run_5_00_energyhivqe_result = {
"1.75": -2373.681781,
"2.00": -2373.694128,
"5.00": -2373.637807,
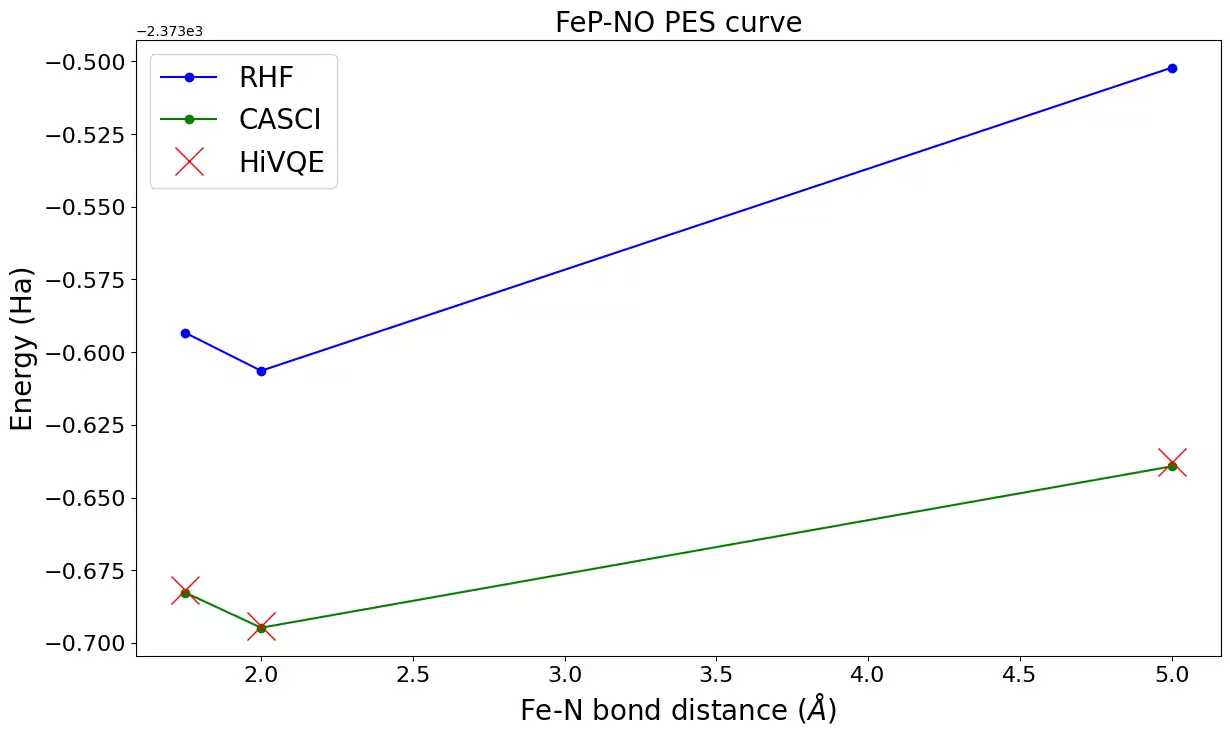
}Step 4: Post-process and compare with classical methods
Classical reference calculation (CASCI-DMRG, maxM=800) results are provided for (22o,22e) to validate HiVQE results.
rhf_result = {
"1.75": -2373.59331683504,
"2.00": -2373.60640773065,
"5.00": -2373.50214278007,
}
casci_result = {"1.75": -2373.6827, "2.00": -2373.6948, "5.00": -2373.6393}fig, ax = plt.subplots(1, 1)
hf_energy = [v for key, v in rhf_result.items()]
casci_energy = [v for key, v in casci_result.items()]
hivqe_energy = [v for key, v in hivqe_result.items()]
distance_ref = [float(key) for key, v in rhf_result.items()]
distance = [float(key) for key, v in hivqe_result.items()]
ax.plot(distance_ref, hf_energy, "-o", label="RHF", c="blue")
ax.plot(distance_ref, casci_energy, "-o", label="CASCI", c="green")
ax.plot(distance, hivqe_energy, "x", label="HiVQE", c="red", markersize=20)
ax.legend(fontsize=20)
ax.tick_params("both", labelsize=16)
ax.set_xlabel("Fe-N bond distance ($\AA$)", size=20)
ax.set_ylabel("Energy (Ha)", size=20)
ax.set_title("FeP-NO PES curve", size=20)
fig.set_size_inches(14, 8)Output:
Tutorial survey
Please take one minute to provide feedback on this tutorial. Your insights will help us improve our content offerings and user experience.